Publications
Selected publications (link to the complete list of our publications)
Kwak Y, Daly CWP, Fogarty EA, Grimson A, Kwak H. (2022) Dynamic and widespread control of poly(A) tail length during macrophage activation. RNA 28(7):947-971.
Kwak Y, Kwak H. (2022) Genome-Wide Identification of Polyadenylation Dynamics with TED-seq. Methods Mol Biol. 2404:281-298.
Kristjánsdóttir K, Dziubek A, Kang HM, Kwak H. (2020) Population-scale study of eRNA transcription reveals bipartite functional enhancer architecture. Nat Commun. 11(1):5963.
Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR (2019) m6A enhances the phase separation potential of mRNA. Nature. 571(7765):424-428. doi: 10.1038/s41586-019-1374-1.
Woo YM, Kwak Y, Namkoong S, Kristjánsdóttir K, Lee SH, Lee JH and Kwak H. (2018) TED-seq identifies the dynamics of poly(A) length during ER stress. Cell Rep. 24(13):3630-3641
Johnson MB, Sun X, Kodani A, Borges-Monroy R, Girskis KM, Ryu SC, Wang PP, Patel K, Gonzalez DM, Woo YM, Yan Z, Liang B, Smith RS, Chatterjee M, Coman D, Papademetris X, Staib LH, Hyder F, Mandeville JB, Grant PE, Im K, Kwak H, Engelhardt JF, Walsh CA, Bae BI. (2018) Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature. 556(7701):370-375.
Chu T, Rice E, Booth GT, Salamanca HH, Wang Z, Core LJ, Longo SL, Corona RJ, Chin LS, Lis JT, Kwak H*, Danko CG*. (in press) Chromatin run-on reveals nascent RNAs that differentiate normal and malignant brain tissue. Nat. Genet. (*co-correspondence)
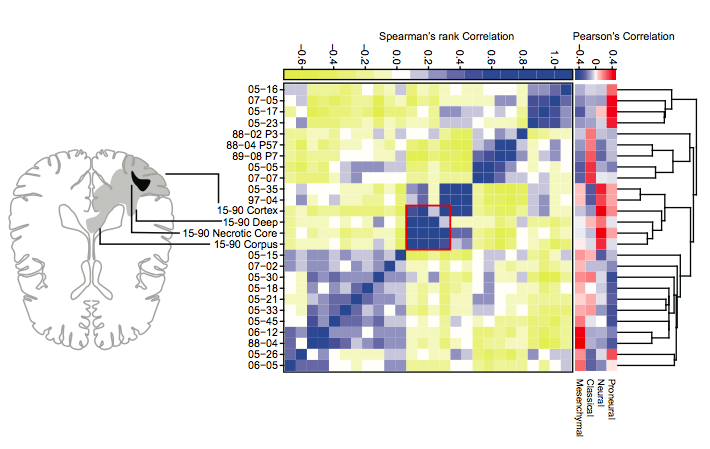 Chromatin Run-On sequencing (ChRO-seq) is a solid tissue compatible version of PRO-seq that can readily be applied to clinical samples. The transcriptional landscape of the brain tumor from human patient samples obtained by ChRO-seq reveals gene expression and the activities of regulatory regions by detecting non-coding enhancer RNAs. We identified tumor associated enhancers that regulate genes characteristic of each molecular subtype of the gliobastoma, and transcription factors that drive the tumor specific transcription programs, showing potential association with clinical outcomes.
Chromatin Run-On sequencing (ChRO-seq) is a solid tissue compatible version of PRO-seq that can readily be applied to clinical samples. The transcriptional landscape of the brain tumor from human patient samples obtained by ChRO-seq reveals gene expression and the activities of regulatory regions by detecting non-coding enhancer RNAs. We identified tumor associated enhancers that regulate genes characteristic of each molecular subtype of the gliobastoma, and transcription factors that drive the tumor specific transcription programs, showing potential association with clinical outcomes.
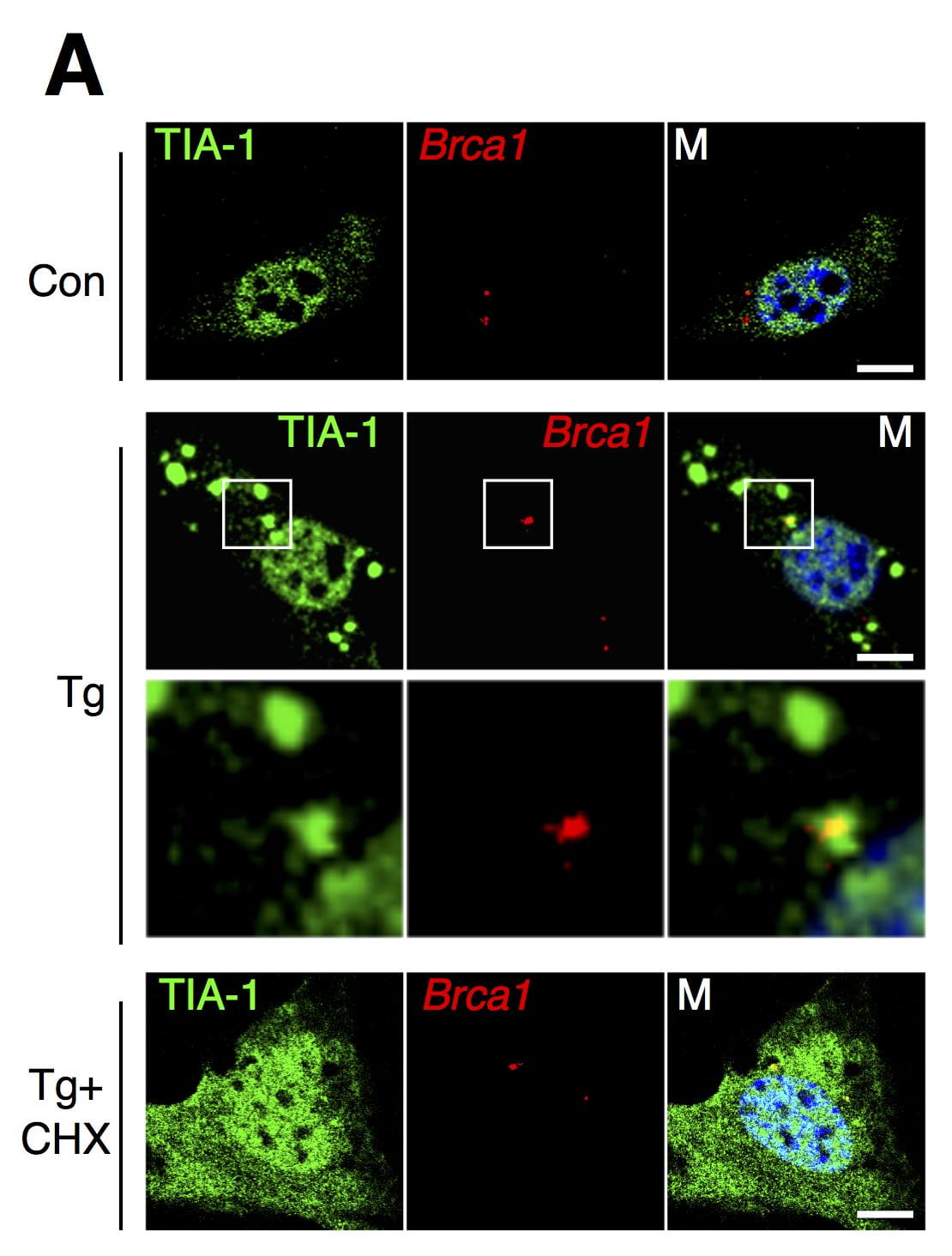
Namkoong S, Ho A, Woo YM, Kwak H*, Lee JH*. (2018) Systematic Characterization of Stress-Induced RNA Granulation. Mol. Cell. (*co-correspondence)
In cell stress, cytoplasmic RNA are known to be sequestered to RNA granules. We investigated whether there is transcript specificity in this stress induced RNA granulation. We used subcellar fractionation, RNA sequencing, and RNA FISH to discover that certain mRNAs (e.g. Brca1) are more prone to relocalize in RNA granules. These transcripts tend to be longer, have more AU-rich elements, and are enriched with genes that affect cell survival and apoptosis.
Jonkers I*, Kwak H*, Lis JT. (2014) Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 3:e02407. (* equal contribution)
We analyzed how fast the RNA polymerase moves along the gene to synthesize RNA. This speed can be a critical checkpoint in controlling the amount of transcription and co-transcriptional RNA processing, but has only been measured in a small number of genes. Here, we used a chemical inhibitor of P-TEFb kinase that prevents RNA polymerase escape from the promoter to visualize the clearance of RNA polymerase along the gene. By analyzing the the time-course of this clearance, we measured the elongation rates of up to a thousand genes at once.
Buckley MS*, Kwak H*, Zipfel WR, Lis JT. (2014) Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights to its regulation. Genes Dev. 28(1):14-19. (* equal contribution)
We showed that the RNA polymerase at the promoter are stably paused, although they are not infinitely stable and can be terminated. This termination was a consequence of increased turnover of RNA polymerase at the promoter rather than a control mechanism during heat-shock response in a major Drosophila heat-shock gene Hsp70.
Kwak H, Lis JT. (2013) Control of transcriptional elongation. Ann Rev Genet. 47:483-508.
This is a broad-spectrum review paper on the mechanisms of RNA polymerase elongation, and the factors affecting this process of productive RNA synthesis.
Kwak H, Fuda NJ, Core LJ, Lis JT. (2013) Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 339(6122):950-3.
This paper demonstrates PRO-seq and PRO-cap methods to map genome-wide RNA polymerases and their start sites in base-pair resolution. We identified different modes of RNA polymerase pausing, and showed how promoter DNA sequences direct early transcription in Drosophila cells.
