Research
Regulatory regions of the genome
The human genome has 3 billion base pairs of DNA, but only a small fraction contains protein coding information. Most other regions have unknown roles (perhaps in maintaining the structure of the genome), but significant fractions are the regulatory regions directing how genetic information is carried through the stages of gene expression.
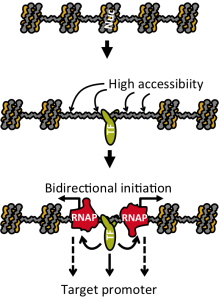
We are focusing on two hallmarks of the regulatory regions.
- They have high chromatin accessibility.
- They can direct transcription at their target genes, but also produce RNA from themselves, called enhancer RNAs (eRNA).
We are using combinations of DNase hypersensitivity, transposon tagmentation assay, and nascent RNA sequencing methods to measure these features and characterize the regulatory regions, especially the mechanisms of enhancer activation.
Research focuses
- How the genetic variation in human population impact gene expression: hierarchical linkages between genetic variation to chromatin accessibility, enhancer activity, and their target gene expression (see Kristjánsdóttir et al, Nat Commun. 2020)
- Identifying the regulatory regions of the cancer genome and exploring the mechanisms of non-coding driver mutations (see Chu et al, Nat Genet. 2018)
New directions
- Direct analysis of eRNA in human peripheral immune cells using human whole blood samples to analyze population and disease traits.
- Test our eRNA TSS (transcription start site) hypothesis using large scale CRISPR prime editing.
